Parallel Serverless Functions at Scale#
Data |
Local |
Coiled |
Speed Up |
|---|---|---|---|
500 GB NetCDF |
305 mins |
4.7 mins |
65x |
The cloud offers amazing scale, but it can be difficult for Python data developers to use. This post walks through how to use Coiled Functions to run your existing code in parallel on the cloud with minimal code changes.
Example: Processing Many Parquet Files on S3#
In the code snippet below we load NYC Taxi Data
from AWS (data lives in region us-east-1), process that data, and then upload the processed data back to AWS.
This type of data processing / cleaning is fairly common in data engineering / science workloads.
import coiled
import pandas as pd
import s3fs
from dask.distributed import print
# Collect files to process
s3 = s3fs.S3FileSystem()
filenames = s3.ls("s3://nyc-tlc/trip data/") # Data is in us-east-1
filenames = [
f"s3://{fn}" for fn in filenames
if "yellow_tripdata_201" in fn # Data files from the 2010s
or "yellow_tripdata_202" in fn # Data files from the 2020s
]
# Define our processing function
def process(filename):
print(f"Processing {filename}")
df = pd.read_parquet(filename)
df = df[df.tip_amount != 0]
outfile = "s3://oss-shared-scratch/jrbourbeau/" + filename.split("/")[-1]
df.to_parquet(outfile)
return outfile
# Process files
print(f"Processing {len(filenames)} files")
results = map(process, filenames)
for outfile in results:
print(f"Finished {outfile}")
print("Done!")
Running this code on my laptop (M1 MacBook Pro) takes a little over five hours.
This is slow for a couple of reasons:
We spend time transferring data from AWS to my laptop and back.
Data files process sequentially, one at a time.
Let’s use Coiled Functions to address both these issues.
Run Functions on the Cloud#
To run the process function on the cloud, we lightly annotate it
with the @coiled.function decorator. Now process automatically runs on a VM in
AWS and returns the result locally.
@coiled.function(
region="us-east-1", # Same region as data
vm_type="m6i.xlarge", # Enough memory to process files
)
def process(filename):
# Everything inside the function stays the same
...
Note that we set region="us-east-1" and vm_type="m6i.xlarge" to make sure the VM the process function
runs on is in the same region as our data and has enough memory to process the data files
(m6i.xlarge instances have 16 GiB of memory).
Adding the @coiled.function decorator reduces the overall runtime to 35.9 minutes
(8.5x faster than the original version that ran locally). This is a nice performance improvement,
but we can do even better by scaling out and running in parallel.
Run in Parallel#
By default @coiled.function decorated functions will run in the cloud, serially, and return the results
back to your local machine. When we want to apply our function many times across different inputs,
we can use the Coiled Funtion .map method to run our function in parallel across inputs on multiple VMs.
In this example, we swap out Python’s builtin map function
for, process.map (everything else stays the same):
# results = map(process, filenames) # Replace this
results = process.map(filenames) # With this
Now the number of VMs used to run process calls will adaptively scale up and down depending on the workload
(see Adaptive Deployments in the Dask docs
for more information), with each VM running a separate process call across inputs.
Running the process function in parallel, we briefly scale up to 33 VMs, and the overall runtime
dropped to 4.7 minutes (65x faster than the original version that ran locally).
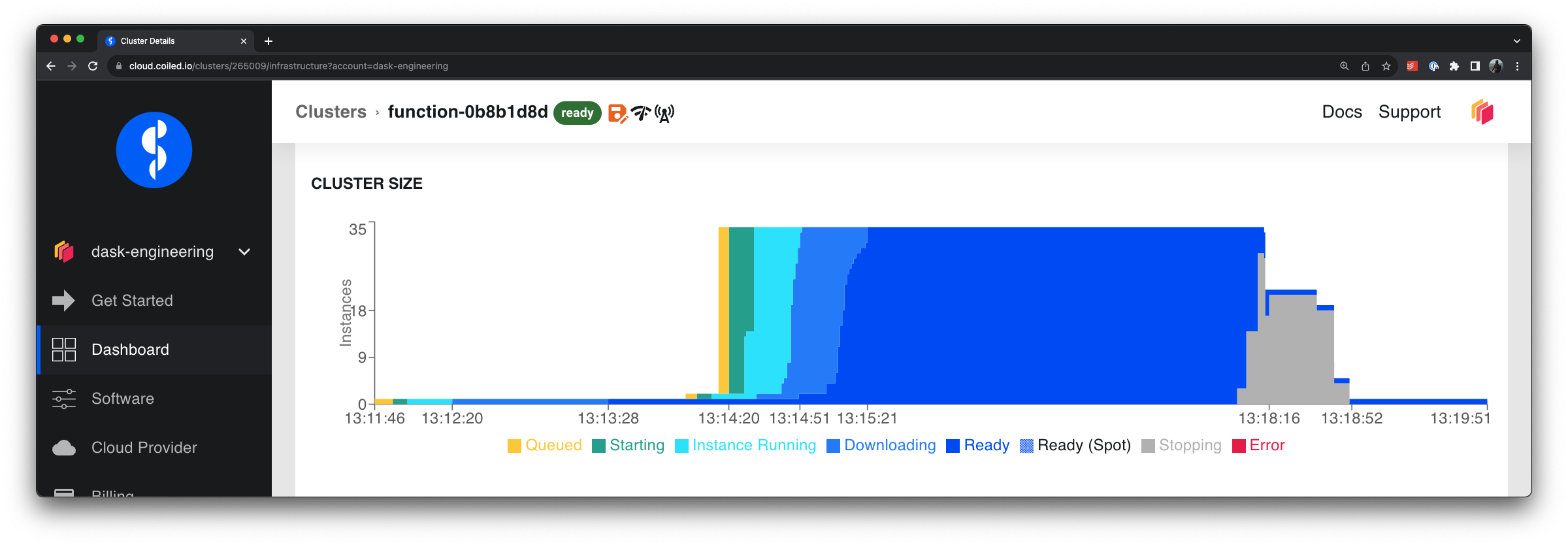
VMs adaptively scaling up to process files quickly in parallel and then scaling down after all files are processed#
Summary#
In this post we demonstrated how to take existing Python code that runs locally and run it in parallel on the cloud with Coiled Functions. With minimal code changes, we were able to improve performance by a factor of 65x, without needing to maintain any additional cloud infrastructure. This makes developers’ lives a little easier and lowers the bar for transitioning code to run on the cloud.
Comparing code runtime between a laptop, single cloud VM, and multiple cloud VMs in parallel#
Full example
import coiled
import pandas as pd
import s3fs
from dask.distributed import print
# Collect files to process
s3 = s3fs.S3FileSystem()
filenames = s3.ls("s3://nyc-tlc/trip data/") # Data is in us-east-1
filenames = [
f"s3://{fn}" for fn in filenames
if "yellow_tripdata_201" in fn # Data files from the 2010s
or "yellow_tripdata_202" in fn # Data files from the 2020s
]
# Define our processing function
@coiled.function(
region="us-east-1", # Same region as data
vm_type="m6i.xlarge", # Enough memory to process files
)
def process(filename):
print(f"Processing {filename}")
df = pd.read_parquet(filename)
df = df[df.tip_amount != 0]
outfile = "s3://oss-shared-scratch/jrbourbeau/" + filename.split("/")[-1]
df.to_parquet(outfile)
return outfile
# Process files
print(f"Processing {len(filenames)} files")
results = process.map(filenames)
for outfile in results:
print(f"Finished {outfile}")
print("Done!")
Want to run this example yourself?
Get started with Coiled for free at coiled.io/start. This example runs comfortably within the free tier.
Copy and paste the code snippet above.