Data-proximate Computing with Coiled Functions#
Coiled Functions make it easy to improve performance and reduce costs by moving your computations next to your cloud data.
It’s common practice for data scientists and researchers to analyze data on their local work computer (often a laptop). This works great for data that’s stored locally on their machine. However, increasingly data is moving to cloud storage system like AWS S3 and Google Cloud Storage. When analyzing cloud data on a laptop, users end up needing to transfer their data from cloud storage to their laptop, which is both slow and expensive.
This post walks through how to use Coiled Functions to move your data-intensive computations to the cloud, alongside your data, to reduce costs and improve performance.
Example: Analyzing NYC taxi data#
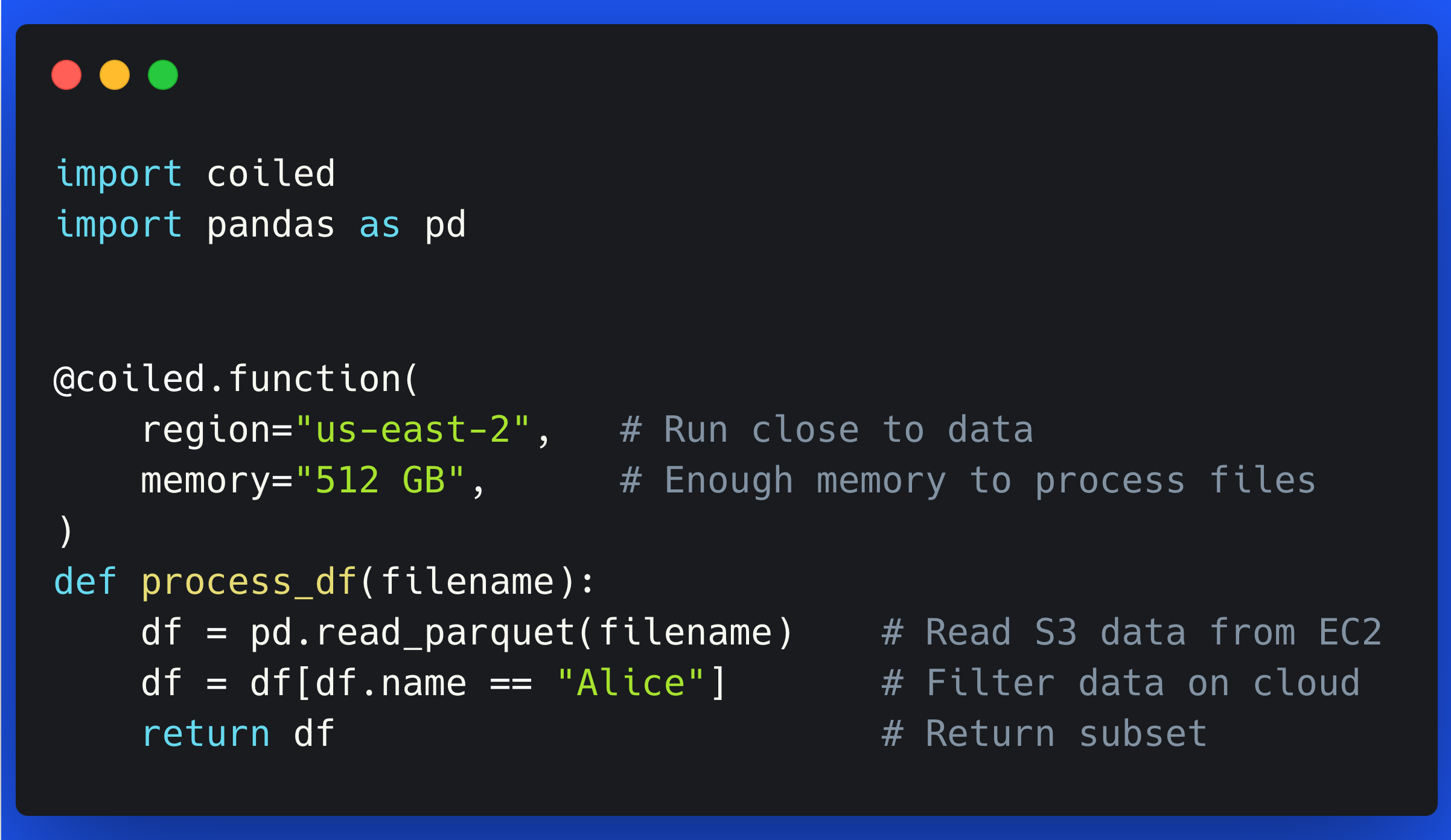
In the code snippet below we load NYC Taxi Data from AWS (data lives in us-east-1), process that data, and then upload the processed version of the data back to AWS. This type of data processing / cleaning is fairly common in data science workloads.
import time
import coiled
import pandas as pd
import s3fs
from dask.distributed import print
t_start = time.time() # Get starting time
# Collect files to process
s3 = s3fs.S3FileSystem()
filenames = s3.ls("s3://nyc-tlc/trip data/") # Data is in us-east-1
filenames = [
f"s3://{fn}" for fn in filenames
if "yellow_tripdata_2022" in fn
]
# Define our processing function
def process(filename):
df = pd.read_parquet(filename)
df = df[df.tip_amount != 0]
outfile = "s3://oss-shared-scratch/jrbourbeau/" + filename.split("/")[-1]
df.to_parquet(outfile)
print(f"Finished {outfile}")
# Process files
print(f"Processing {len(filenames)} files")
for filename in filenames:
print(f"Processing {filename}")
process(filename)
print("Done!")
# Print total runtime
t_end = time.time()
print(f"Took {t_end - t_start / 60:0.2f} minutes")
Running this code on my laptop (M1 MacBook Pro) in Madison, WI takes 11.25 minutes.
In this case, most of the time is spent transferring data from AWS to my laptop and then uploading the processed data on my laptop back to AWS.
Run functions on the cloud#
Given our data is already on AWS, let’s update our process function to also run on the cloud in AWS. We can use Coiled Functions to decorate our existing process function and have it automatically run on a VM in AWS.
@coiled.function(
region="us-east-1", # Same region as data
vm_type="m6i.large", # Enough memory to process files
)
def process(filename):
# Everything inside the function stays the same
...
Note that we set region="us-east-1" and vm_type="m6i.large" to make sure the VM this process function runs on is in the same region as our data and has enough memory to process the data files (m6i.large instances have 8 GiB of memory).
When I add the @coiled.function decorator and rerun the script, it takes 2.82 minutes to run (note that includes the startup time for the cloud VM), which is about a 4x overall speedup. We also never had to transfer data outside of AWS, so we didn’t need to worry about data transfer costs.
Summary#
Moving computations that process cloud-based data to be co-located alongside the data can both improve code runtime performance and reduce data transfer costs.
Coiled Functions is a simple API for moving existing Python functions to run on the cloud with minimal code changes. This makes developers’ lives a little easier and lowers the bar for transitioning code to run on the cloud.
Learn more#
Check out the Coiled Functions documentation.
See the “How to Train a Neural Network on a GPU in the Cloud with Coiled Functions” blogpost from Patrick Hoefler for another handy use case for
@coiled.function.We highly value user feedback. If you try Coiled Functions, feel free to let us know about your experience in the coiled/feedback GitHub repo or by emailing support@coiled.io.
Get started with Coiled for free at coiled.io/start.